CRISPR is a eukaryote genome-editing tool that is based on endogenously occurring bacterial defense systems. CRISPR technology has been extensively applied in basic science and clinical research since its initial development and characterization in 2013 (1, 2, 3).
CRISPR gene editing requires three components: (1) a guide RNA (gRNA), (2) a Cas9 endonuclease, and (3) a protospacer adjacent motif (PAM). The gRNA are small single stranded RNA molecules that hold two distinct regions. The first gRNA region is a gene targeting region that is designed to complement a 15-24 base pair segment of the gene of interest’s DNA sequence. The second gRNA region is a scaffold. The gRNA scaffold region binds to a Cas9 endonuclease while leaving the host DNA compliment region of the gRNA unbound. The Cas9-gRNA complex may then scan the host cell’s genomic DNA for a specific 2-6 base pair sequence referred to as the PAM. Following PAM recognition, the gene targeting region of the gRNA may anneal to the host DNA adjacent to the PAM sequence. Annealing of the gRNA target region then triggers Cas9 cleavage of the host DNA adjacent to the PAM sequence.
Generation of the SpCas9 knock-in mouse, along with the development of the short SaCas9 and NmCas9 variants, have opened several applications for AAV transgene delivery systems in CRISPR experiments. (4, 5). As a result of these advancements, AAVs have become the first choice for CRISPR transgene delivery due to their reliability, versatility, and safty (6, 7).
SpCas9
CRISPR gene knockout experiments can use AAVs for transgene delivery through several distinct approaches. First, the duel-vector approach uses simultaneous infection with two separate AAVs. In this approach, one AAV is designed to drive the expression of SpCas9 with a second AAV to drive the expression of a sgRNA. While well established and functional, the duel-vector approach requires that each cell is successfully infected with both viruses and that both viruses successfully drive expression of their genetic payloads. Variability in the infection or expression rates of each AAV may therefore result in less than ideal knockout coverage.
As an alternative, Zhang’s group from the Broad Institute at MIT bread two lines of SpCas9 knock-in mice. One mouse line displays ubiquitous expression of SpCas9 and a second displays Cre-dependent constitutive expression of SpCas9. This group then and demonstrated that utilizing AAV to deliver sgRNA can precisely induce tumorigenic mutations in the KRAS gene (4). This experiment clearly shows the utility and flexibility of using AAV in CRISPR gene editing experiments, and these SpCas9 mouse lines are now available from the Jackson Lab.
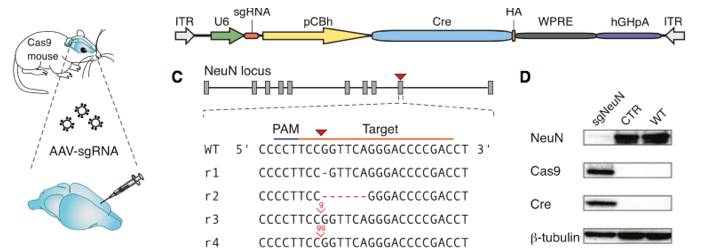
Constitutive genome editing in the brains of Cre-dependent Cas9 mice. An AAV carrying gRNA targeting the neuronal protein NeuN and Cre was delivered by stereotaxic injection into the brain of the Cre-dependent Cas9 mouse. Illustrations depict a schematic of the AAV vector used for these experiments, and well as the sgRNA design for targeting CRISPR edits to NeuN. The representative immunoblot demonstrates a NeuN knockout in Cas9 expressing mice. The bar graphs below demonstrate a significant reduction in NeuN expression following AAV-based sgRNA delivery. Figure from reference 4.
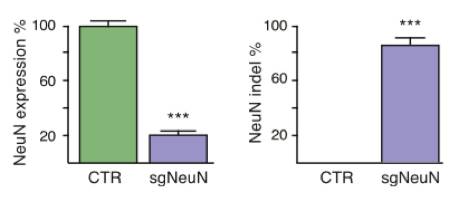
Figures from Ref. 5
Schematic map of systemic genome editing resulting in tumorigenesis. A schematic representation of AAV delivery into Cre-dependent Cas9 mice as well as a diagram of the experimental workflow. Images demonstrate tumorigenesis following intrathecal delivery of AAV designed to express Cre and gRNA targeting the Kras, p53, and Lkb1 genes. Shown below is a schematic representation of the AAV vector used to induce gRNA and Cre expression.
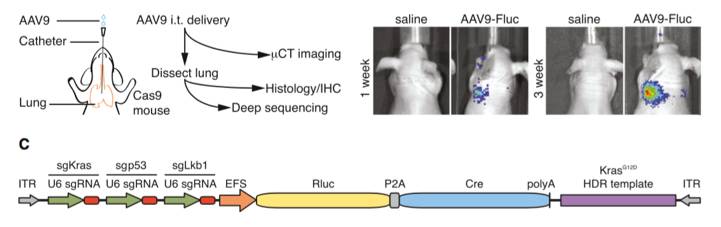
Konermann’s group from MIT discovered that a short 15-bp sgRNA can arrest a wild-type SpCas9 at the target host genome locus without cutting the host DNA (11). Short arresting sgRNA sequences can be paired with a modified gRNA scaffold that includes two MS2 loops and is capable of recruiting a dRNA transcription activation complex. The net effect of these gRNA modifications is the assembly of a SpCas9 complex that can be targeted toward the transcription start region of a gene to drive transcription activation. This technology thus provides a mechanism for gene overexpression that circumvents base pair size limitations and thus allows for overexpression of large and complex genes. Zhang et al. found that similar gRNA modifications can be used to drive gene expression with SaCas9 (12).
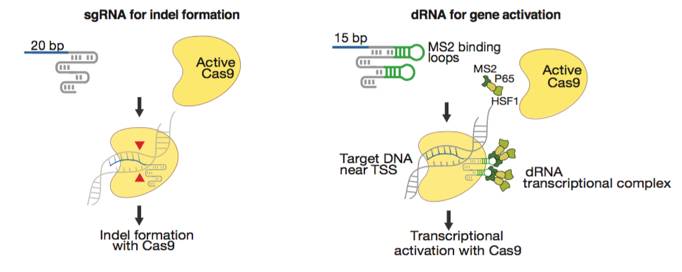
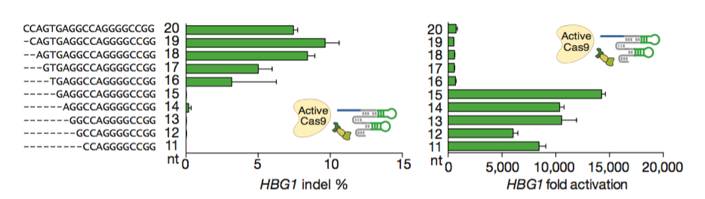
Figures from Ref. 11
Gene activation by active Cas9 paired with a short arresting sgRNA-MS2. Schematic diagrams of active catalytic Cas9 paired with traditional sgRNA for DNA cleavage or paired with a short arresting sgRNA-MS2 for transcription activation. Bar Graphs demonstrate that sgRNA-MS2 with guides of >15nt produce cleavage but produce transcription activation with guides of ≤15nt.
SaCas9
Recombinant AAVs can incorporate transgenes up to a maximum size of 5kb. Incorporation of the 4.3kb SpCas9 into an rAAV leaves little room for additional DNA elements such as cell-type specific promoters or guide RNA expression cassettes. Thus, the utility of SpCas9-rAAV is limited by the size of SpCas9. To subvert this limitation Zhang’s group characterized the smaller 3.3kb Cas9 orthologue SaCas9. They then demonstrated that SaCas9 can edit the genome with efficiencies similar to SpCas9 and can be functionally incorporated into rAAV with >1kb for available for the inclusion of additional DNA elements. Further, this study demonstrated that both SaCas9 and a guide RNA expression cassette can be incorporated into a single AAV vector and showed significant knockout of the liver gene Pcsk9 using this single AAV approach.
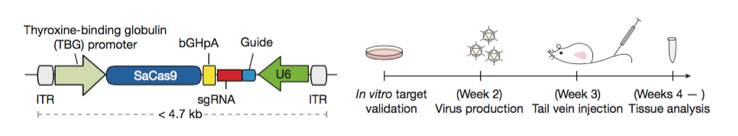
Single AAV vector CRISPR genome editing system. A schematic diagram of the single AAV CRISPR vector (Left). A Schematic diagram of the experimental design used to demonstrate in vivo knockout efficacy of the single AAV single AAV CRISPR vector (right). Figure from reference 5.
The development of AAV-SaCas9 vectors reduces the need for SpCas9-mouse lines, and may thus result in substantial time and cost savings with the subsequent reduction in crossbreeding and mousseline genetic testing costs. This approach has since been well adopted and applied extensively across both basic science and experimental therapeutic research. PackGene offers a set of optimized entry plasmids for AAV-SaCas9 applications.
Refferences
2) Le Cong et al. Science 339,819 (2013); DOI: 10.1126/science.1231143
3) Prashant Mali et al. Science 339,823 (2013); DOI: 10.1126/science.1232033
4) Randall J. Platt et al. Cell 159, 1–16 (2014); DOI: 10.1016/j.cell.2014.09.014
5) F. Ann Ran et al. Nature 520, 186 (2015); doi:10.1038/nature14299
6) Florian Schmidt and Dirk Grimm. Biotechnology J. 10, 258 (2015); DOI0.1002/biot.201400529
7) David B. T. Cox et al. Nature Medicine 21,121 (2015); doi: 10) 1038/nm.3793.
8) C. Long et al., Science 10.1126/science.aad5725 (2015)
9) C. E. Nelson et al., Science 10.1126/science.aad5143 (2015)
10) M. Tabebordbar et al., Science 10.1126/science.aad5177 (2015).
11) Dahlman JE et al., Nat Biotechnol. 2015 Nov;33(11):1159-61.
12) Nishimasu et al., Cell 162, 1113–1126 (2015)
